Credit: Cell (2023). DOI: 10.1016/j.cell.2023.08.029
Research led by multiple institutions in China has examined how small, furry, viral vectors affect the spread and evolution of viruses. They report the identification of 669 viruses, including 534 novel viruses, greatly expanding our knowledge of the mammalian virome, including previously unknown coronaviruses and orthorubulaviruses.
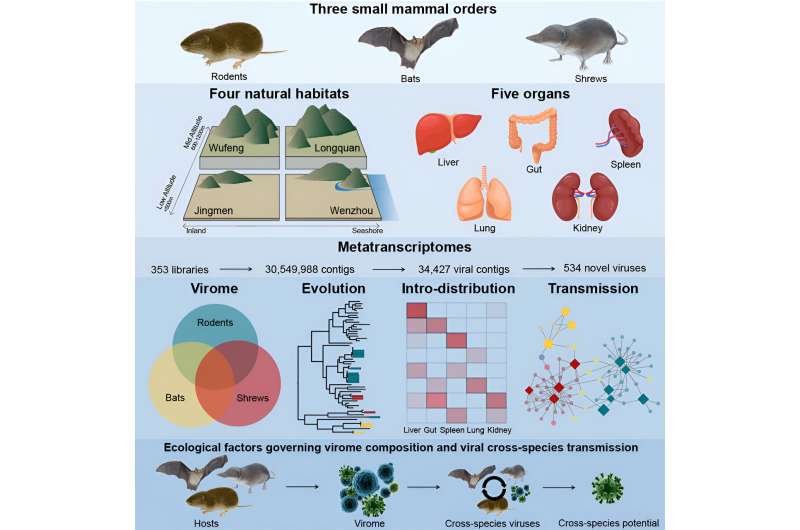
In their paper, “Host traits shape virome composition and virus transmission in wild small mammals,” published in Cell, the team used meta-transcriptomic sequencing of internal organ and fecal samples from 2,443 wild bats, rodents, and shrews from four Chinese habitats. Viruses were identified in nearly all animals studied.
Researchers identified viruses related to known human or domestic animal pathogens (e.g., Rotavirus A, Seoul virus, Wenzhou mammarenavirus), including coronaviruses, which have caused diseases like SARS and COVID-19. There were also newly identified viruses from the orthorubulaviruses, porcine epidemic diarrhea virus (PEDV), swine acute diarrhea syndrome coronaviruses (SADS-CoVs), and SARS-related coronaviruses.
The study provided robust evidence of cross-species virus transmission. For instance, viruses were found in multiple species of wild small mammals, indicating that these viruses can move between different animal species, potentially including humans.
The identification of a bat arenavirus suggests that arenaviruses have made host jumps from rodents to bats. This finding provides phylogenetic insights into the evolution of these viruses.
In some virus families, Astroviridae, Paramyxoviridae, and Picornaviridae, the study found versions that were relatively distinct from known sequences.
Bats, rodents, and shrews were compared in terms of virus richness. Shrews were found to harbor the most viruses in total and the most found in a single species, with up to 150 viruses from 29 viral clades identified in Smith’s shrews. Some of these shrew viruses were related to those previously identified in invertebrates, while others were distinct.
The team observed cross-species virus transmission at taxonomic levels across species, genus, family, and order, where the same virus was found in three or more mammalian species. Viruses with multi-organ distributions within hosts were more likely to be found in other host species. This suggests that if a virus is present in multiple internal organs, it has a higher chance of spreading to other species.
Bats are often considered to harbor more viruses than rodents. Of the animals sampled in the study, bats did have the highest richness, followed by rodents and shrews. While the total number of viruses identified in bats was more than that in rodents, the average number of viruses identified per species of bats and rodents was similar.
More information:
Yan-Mei Chen et al, Host traits shape virome composition and virus transmission in wild small mammals, Cell (2023). DOI: 10.1016/j.cell.2023.08.029
Journal information:
Cell
© 2023 Science X Network
Citation:
Cross-species virus transmission found in several species of small furry animals (2023, September 25)
retrieved 25 September 2023
from https://phys.org/news/2023-09-cross-species-virus-transmission-species-small.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
>>> Read full article>>>
Copyright for syndicated content belongs to the linked Source : Phys.org – https://phys.org/news/2023-09-cross-species-virus-transmission-species-small.html










