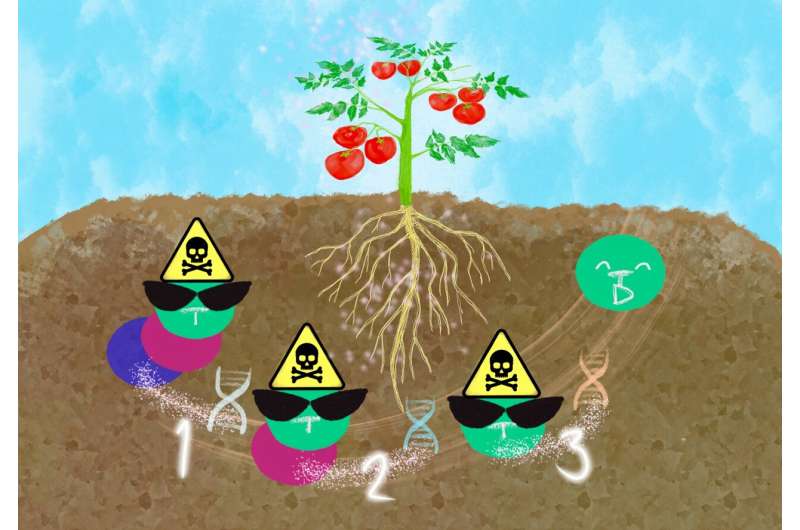
Four corresponding genes SpGH3-4, SpGH39-1, SpGH3-1, and SpGH3-3 jointly work to hydrolyze tomatine to tomatidine in the tomato rhizosphere. Credit: KyotoU Global Comms/Jake Tobiyama
Regardless of how one says “tomato,” they all contain tomatine, a toxin in the plant’s green fruit, leaves, and roots. Tomatoes produce the bitter-tasting compound—a major plant-specialized metabolite secreted from the roots—to defend against pathogens and foragers.
Such metabolites function as nutrients and chemical signals, affecting the formation of microbial communities that greatly influence plant growth.
Previous studies have found that plant-based organic toxins—saponins, such as tomatine—alter the microbial community around tomato roots by increasing the bacterium Sphingobium. Yet, what remained unknown was how the microbe’s colonies in the tomato rhizosphere—the soil surrounding the roots—dealt with tomatine.
Now, a research group led by Kyoto University has revealed that Sphingobium possesses a series of enzymes that hydrolyze tomatine, detoxifying it.
“We also identified enzymes that convert the steroidal tomatidine to non-toxic, smaller compounds,” says Akifumi Sugiyama of KyotoU’s Research Institute for Sustainable Humanosphere.
“Our discovery of these metabolites helps us to understand how soil microorganisms cope with plant-derived toxic compounds to inhabit the rhizosphere successfully,” adds Masaru Nakayasu, also at RISH.
Sugiyama’s team isolated several bacteria from tomato roots and tomatine-infused soil and identified the bacterial strain RC1, which downgrades tomatine and utilizes it as a carbon source.
Sequence analyses of RC1’s genes demonstrated that the expression of several genes of the glycohydrolase family increased in the presence of tomatine. The expressions of the proteins encoded by the genes of the bacterium E Coli confirmed their ability to degrade tomatine in vitro.
“We had thought the four sugars attached to tomatine degraded in some order but discovered that the four corresponding genes SpGH3-4, SpGH39-1, SpGH3-1, and SpGH3-3 jointly work to hydrolyze tomatine to tomatidine,” noted Kyoko Takamatsu at KyotoU’s Graduate School of Agricultural Science.
The authors anticipate further efforts to develop saponins other than tomatine and analyze how the saponin-degrading genes affect the interaction between plants and bacterial communities in the rhizosphere.
“Given that many plant-specialized metabolites offer human health benefits, we can engineer the bacterial genes based on their enzymatic functions to produce new bioactive compounds for human applications,” concluded Sugiyama.
The study is published in the journal mBio.
More information:
Masaru Nakayasu et al, Tomato root-associated Sphingobium harbors genes for catabolizing toxic steroidal glycoalkaloids, mBio (2023). DOI: 10.1128/mbio.00599-23
Journal information:
mBio
Citation:
Examining the toxins in the common tomato (2023, October 6)
retrieved 6 October 2023
from https://phys.org/news/2023-10-toxins-common-tomato.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
>>> Read full article>>>
Copyright for syndicated content belongs to the linked Source : Phys.org – https://phys.org/news/2023-10-toxins-common-tomato.html










