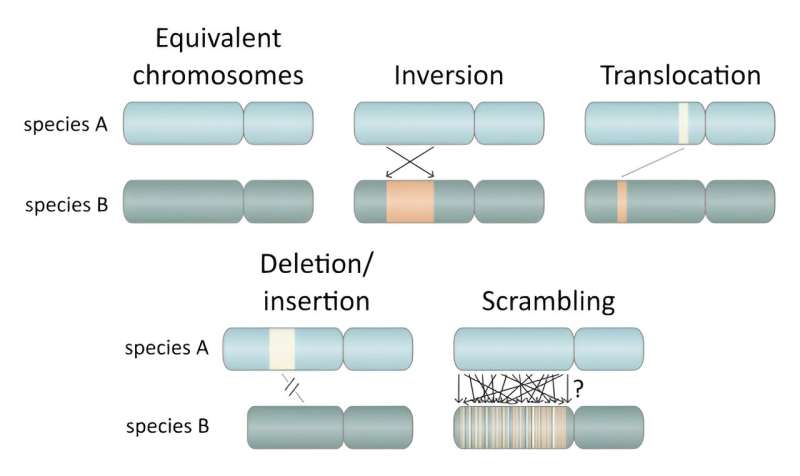
Some of the common types of genomic rearrangements. These rearrangements form the basis of evolution, as they give way for new combinations of genes that could result in new traits that allow species to better adapt to their environment. Credit: Michael J. Mansfield (OIST).
When two animals look the same, eat the same, behave the same way, and live in similar environments, one might expect that they belong to the same species.
However, a tiny zooplankton skimming the ocean surfaces of microscopic food particles challenges this assumption. Researchers from Osaka University, University of Barcelona and the Okinawa Institute of Science and Technology (OIST) have analyzed the genome of Oikopleura dioica from the Seto Inland Sea, the Mediterranean, and the Pacific Ocean around the Okinawa Islands, and in doing so, they have raised numerous questions about speciation and the role of gene location in the genome.
Their results have been published in Genome Research. “Oikopleura is opening new avenues into genomic research,” says Dr. Charles Plessy from the Genomics and Regulatory Systems Unit at OIST and co-first author of the paper.
“As a model animal, it allows us to study the mechanisms for genome changes in the lab as they happen at a very large scale and speed, which is an enormous opportunity.”
Oikopleura dioica is a tiny zooplankton that inhabits the ocean surface worldwide, and which is used as a model organism in developmental biology.
As a chordate, the organism shares key genetic and developmental traits with vertebrates, including the presence of a notochord, which is a chord-like central nerve bundle like a spinal column, but without bones. Moreover, its compact genome, the smallest non-parasitic animal genome reported to date, facilitates large-scale genomic analysis.
The genomic tower of Babel
The researchers worked on three lineages of Oikopleura dioica sampled from three seas around the world, but though the morphological, behavioral, and ecological characteristics of the lineages are virtually the same, the genomes differ massively.
Think of the genome as a shared language between all members of a single species, stored within the nucleus of every cell and containing the complete set of genetic material to make that species. Like how grammar determines the arrangement of words to convey specific meanings, so too are the basic units of information in the genome—the genes—regulated in relation to one another when they’re transcribed and translated into the fundamental building blocks of life, proteins.
Gene regulation involves multiple factors that impact the activation or rate of gene transcription, such as other genes, molecules in the cell, hormones, and many others.
What’s puzzling about the Oikopleura dioica genome is that the languages of the three lineages don’t seem to match, despite them having almost identical physical characteristics. That is, the ‘meaning’ produced by their genes are mostly the same, while the genomic languages are wildly different between them.
The researchers use the term ‘scrambling’ to describe the phenomenon observed in Oikopleura dioica, a term which originates in linguistics to denote a phenomenon whereby sentences are formulated using a variety of different word orders without any change in meaning.
While this phenomenon does not occur in English (but does in Japanese and other languages), an English example would be if the sentence “the genome of Oikopleura dioica is highly scrambled” could be rearranged to “highly scrambled Oikopleura dioica the genome of is” without a change in meaning. While genomic rearrangements are common to all species, and genome scrambling has been observed in a few species over a very long time, Oikopleura dioica surpasses what was previously thought possible.
Ribbon charts comparing chromosomes across species: human vs. house mouse, two species of Ciona sea squirts, and two Oikopleura dioica lineages. Black lines depict chromosomes, lengths measured in megabase pairs (Mb), here used to indicate the coordinates of the genes on the chromosomes. Blue boxes represent matching regions between chromosomes. Orange ribbons denote matches with the same gene order, while blue ribbons indicate reversed order. Credit: Charles Plessy (OIST)
Evolution at breakneck speed
The researchers compared the genetic sequences of the three lineages, which led them to estimate that they shared a common ancestor around 25 million years ago, with the lineages from Barcelona and Osaka being more closely related than the Okinawa lineage, having diverged ~7 million years ago. For comparison, humans diverged from mice 75-90 million years ago.
From their phylogenetic analyses, the researchers estimated the rate of genomic rearrangements for different species as a quantifiable measure for how quickly they evolve. From this, the researchers found that the rate for Oikopleura dioica is more than ten times higher than comparable species of Ciona sea squirts.
As Dr. Michael J. Mansfield from the unit and co-first author on the paper puts it, “The Oikopleura is one of the fastest evolving animals in the world. Animals, especially chordates, don’t normally rearrange their genomes to this extent, at this speed.”
With all this genome scrambling taking place between the Oikopleura dioica lineages, from a genomic perspective it’s mystifying that they can retain such similar characteristics.
“Our results suggest that while genomic organization is important, especially for something as complex as human beings, we should not forget the individual genes,” suggests Dr. Plessy. Studying genes and genomes can offer two different perspectives on the same phenomenon—as Dr. Mansfield explains it, “There are scientists studying anatomy and others who study individual neurons—but both are answering questions about the brain.”
Who’s asking?
Genome scrambling poses important questions about evolution and classifying life into species. On the one hand, the researchers show that even if the three lineages of Oikopleura dioica are virtually identical morphologically and functionally, their genomes are extremely scrambled, which could suggest that they belong to different species, though the researchers stress that their intention is not to classify them here. On the other hand, the scrambled yet analogous gene expression could warn against an overreliance on genomics for classifying species.
Ultimately, however, “species don’t need us. If you remove humans, the animals are the same—it doesn’t matter how we classify them,” as Dr. Plessy phrases it. Instead, the concept of species is fluid, depending on whether it’s for conservation purposes, for legislation, as a microbiologist or a zoologist, or whatever the reason is. “The question ‘what is a species?’ can be answered with another question: why do you ask?”
For Dr. Plessy, Dr. Mansfield, and their collaborators around the world, this paper is the culmination of a long process of cultivating different lineages of Oikopleura dioica and developing bioinformatic tools capable of analyzing their chaotic genomes. Professor Nicholas Luscombe, head of the unit at OIST, is optimistic about the research potential of the study and the animals.
“We initially assumed that all Oikopleura would have similar genomes, but we were amazed to see such huge differences with so much scrambling between them. We want to use Oikopleura to learn more about the nature of genomic rearrangements.”
This is just the beginning—the researchers are far from done studying the enigmatic zooplankton. “We have already learned so much from the Oikopleura, but we have yet to explore the full extent of the diversity of the species at a global scale,” Dr. Plessy says.
Dr. Mansfield quotes the great biologist Jacques Monod with “what’s true for E. coli is true for the elephant”—with the tools developed for this study, the teams can now turn their attention to other species. “We went into this thinking that all Oikopleura dioica were the same, but we have shown the opposite. How often is that true for other species, and how much more is there to know about the mechanisms of genome scrambling?”
More information:
Charles Plessy et al, Extreme genome scrambling in marine planktonicOikopleura dioicacryptic species, Genome Research (2024). DOI: 10.1101/gr.278295.123
Citation:
Genomic analysis of a species of zooplankton questions assumptions about speciation and gene regulation (2024, April 26)
retrieved 26 April 2024
from https://phys.org/news/2024-04-genomic-analysis-species-zooplankton-assumptions.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
>>> Read full article>>>
Copyright for syndicated content belongs to the linked Source : Phys.org – https://phys.org/news/2024-04-genomic-analysis-species-zooplankton-assumptions.html
