
A study by the University of Göttingen on Mesotaenium endlicherianum, an alga closely related to land plants, revealed crucial genetic insights. By analyzing the alga’s response to various environmental conditions, researchers uncovered shared genetic mechanisms between algae and land plants, deepening understanding of plant evolution and resilience.
A research team from Göttingen University leads an investigation into 10 billion RNA snippets to identify “hub genes.”
The majority of the Earth’s land surface is adorned with a diverse array of plants, which constitute the majority of biomass on land. This remarkable diversity spans from delicate mosses to towering trees. This astounding biodiversity came into existence due to a fateful evolutionary event that happened just once: plant terrestrialization. This describes the point where one group of algae, whose modern descendants can still be studied in the lab, evolved into plants and invaded land around the world.
An international group of researchers, spearheaded by a team from the University of Göttingen, generated large-scale gene expression data to investigate the molecular networks that operate in one of the closest algal relatives of land plants, a humble single-celled alga called Mesotaenium endlicherianum. Their results were published in Nature Plants.

Liquid samples of Mesotaenium endlicherianum in a laboratory flask, which are about to be combined with fresh medium under sterile conditions. Credit: Janine Fürst-Jansen
Unveiling Algal Resilience
Using a strain of Mesotaenium endlicherianum that has been kept safe in the Algal Culture Collection at Göttingen University (SAG) for over 25 years and the unique experimental set-up there, the researchers exposed Mesotaenium endlicherianum to a continuous range of different light intensities and temperatures.
Janine Fürst-Jansen, researcher at the University of Göttingen, states: “Our study began by examining the limits of the alga’s resilience – to both light and temperature. We subjected it to a wide temperature range from 8 °C to 29 °C. We were intrigued when we observed the interplay between a broad temperature and light tolerance based on our in-depth physiological analysis.”
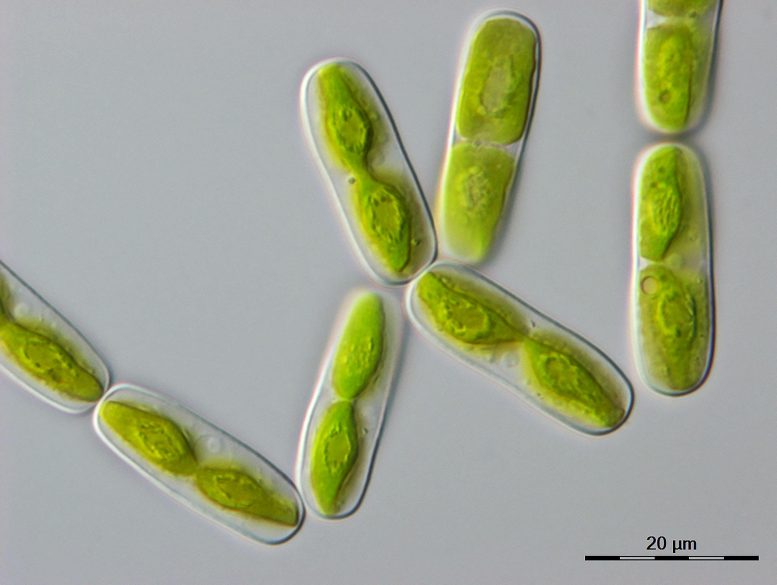
Microscope image of one of the closest algal relatives of land plants, a single-celled alga called Mesotaenium endlicherianum (20 micrometers corresponds to 0.02 millimeters). Credit: Tatyana Darienko
How the algae respond was not only investigated on a morphological and physiological level, but also by reading the information of about 10 billion RNA snippets. The study used network analysis to investigate the shared behavior of almost 20,000 genes simultaneously. In these shared patterns, “hub genes” that play a central role in coordinating gene expression in response to various environmental signals were identified.
This approach not only offered valuable insights into how algal gene expression is regulated in response to different conditions but, combined with evolutionary analyses, how these mechanisms are common to both land plants and their algal relatives.

Samples of Mesotaenium endlicherianum that have been kept safe in the Algal Culture Collection at Göttingen University (SAG) for over 25 years. This image shows the unique experimental set-up that allowed the researchers to expose Mesotaenium endlicherianum to a continuous range of different light intensities and temperatures. Credit: Janine Fürst-Jansen
Discovering Evolutionary Genetic Mechanisms
Professor Jan de Vries, University of Göttingen, says: “What is so unique about the study is that our network analysis can point to entire toolboxes of genetic mechanisms that were not known to operate in these algae. And when we look at these genetic toolboxes, we find that they are shared across more than 600 million years of plant and algal evolution!”
As Armin Dadras, PhD student at the University of Göttingen, explains: “Our analysis allows us to identify which genes collaborate in various plants and algae. It’s like discovering which musical notes consistently harmonize in different songs. This insight helps us uncover long-term evolutionary patterns and reveals how certain essential genetic ‘notes’ have remained consistent across a wide range of plant species, much like timeless melodies that resonate across different music genres.”
Reference: “Environmental gradients reveal stress hubs pre-dating plant terrestrialization” by Armin Dadras, Janine M. R. Fürst-Jansen, Tatyana Darienko, Denis Krone, Patricia Scholz, Siqi Sun, Cornelia Herrfurth, Tim P. Rieseberg, Iker Irisarri, Rasmus Steinkamp, Maike Hansen, Henrik Buschmann, Oliver Valerius, Gerhard H. Braus, Ute Hoecker, Ivo Feussner, Marek Mutwil, Till Ischebeck, Sophie de Vries, Maike Lorenz and Jan de Vries, 28 August 2023, Nature Plants.
DOI: 10.1038/s41477-023-01491-0
>>> Read full article>>>
Copyright for syndicated content belongs to the linked Source : SciTechDaily – https://scitechdaily.com/green-ancestors-decoding-the-secrets-of-600-million-years-of-plant-life/










