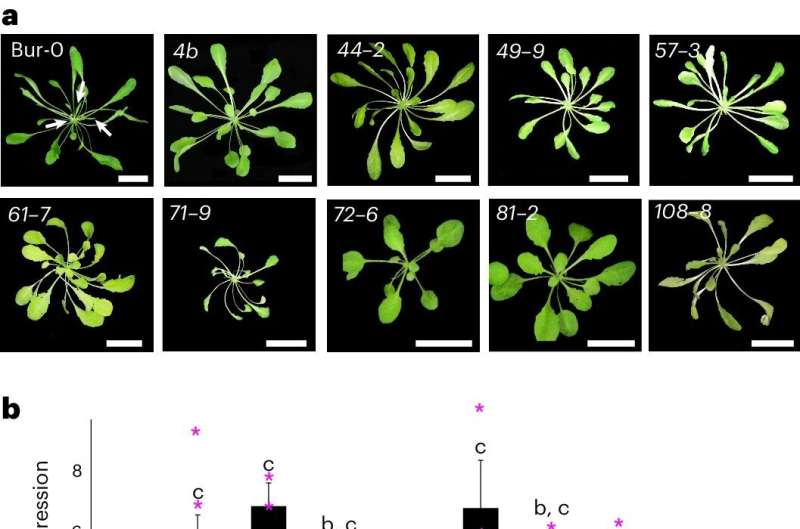
A genetic suppressor screen identifies RdDM to be the major pathway for repeat expansion-induced epigenetic silencing. a, Phenotypes (denoted by their original screen identifiers) of the isolated suppressors compared to Bur-0. The irregularly impaired leaves are marked by white arrows in the Bur-0 wild type. Scale bars, 2 cm. b, Relative IIL1 expression levels in genetic suppressors identified through the genetic screen. The numbers represent the original screen identifiers, and the corresponding genes identified after cloning are shown below. Average expression levels based on three biological replicates for each line (except for Bur-0 and fug1, where n = 5 and 4, respectively) are shown. Asterisks (*) denote individual data points. P values are based on a one-way analysis of variance with Tukey’s post hoc test, and lines with different letters are significantly different from each other (P 0.85) are colored red, and red crosses show the putative causal alleles. Credit: Nature Plants (2024). DOI: 10.1038/s41477-024-01672-5
Monash University biologists have shed light on the intricate molecular mechanisms that are responsible for gene silencing induced by expanded repeats in an international study published today in Nature Plants.
This phenomenon has been linked to a number of hereditary illnesses, including Friedreich’s ataxia in humans, and causes growth abnormalities in plants such as Arabidopsis thaliana.
The research aimed to understand the mechanism by which enlarged repeats cause epigenetic silencing, an essential procedure for controlling gene expression.
Discovering novel components that are necessary for this silencing process was accomplished by the researchers using a plant model that presents the symptoms of growth defects at higher temperatures but not at lower temperatures.
SUMO protease FUG1, histone reader AL3, and chromodomain protein LHP1 were identified as the three most important actors, according to the study.
“These proteins come together to create an essential module required for epigenetic silencing induced by repeat expansion,” said lead study author Dr. Sridevi Sureshkumar, who heads the Genetics at the Core Research Group at the Monash University School of Biological Sciences.
“Our research reveals the crucial role that these proteins play in orchestrating gene silencing that is triggered by expanded repeats,” Dr. Sureshkumar said.
“The awareness of these systems not only contributes to the advancement of our understanding of plant biology but also offers insights into diseases that affect humans,” she said.
During the course of the research, modern genetic screening methods and yeast two-hybrid tests were utilized in order to determine that FUG1, an uncharacterized SUMO protease, is a significant participant in epigenetic silencing. Following further analysis, it was shown that FUG1 interacts with AL3, which is a histone reader that is known to bind to particular histone marks that are related to effective gene expression.
In addition, the researchers found that the AL3 protein interacts with LHP1, which is a chromodomain protein that plays a role in the dissemination of restrictive histone marks. The reversal of gene silencing and the suppression of repeat expansion-associated symptoms were both brought about by the loss of function of any one of these components during the experiment.
“These findings highlight the importance of post-translational modifiers and histone readers in epigenetic regulation,” Dr. Sureshkumar said.
“Our study paves the way for further research into the role of these proteins in various biological processes and human diseases,” she said.
“The findings not only present potential consequences for human health but also contribute to our understanding of plant biology, which is already advanced.”
Dr. Sureshkumar, who led this international study involving Institutions in the UK, China, Canada, India, and Australia, said that multinational collaboration helped them make progress across diverse aspects of this research.
Dr. Sureshkumar said this research could potentially be a pathway for the development of novel therapeutic techniques that target epigenetic dysregulation in people who suffer from hereditary illnesses.
More information:
Sridevi Sureshkumar et al, SUMO protease FUG1, histone reader AL3 and chromodomain protein LHP1 are integral to repeat expansion-induced gene silencing in Arabidopsis thaliana, Nature Plants (2024). DOI: 10.1038/s41477-024-01672-5
Citation:
Uncovering key players in gene silencing: Insights into plant growth and human diseases (2024, April 19)
retrieved 19 April 2024
from https://phys.org/news/2024-04-uncovering-key-players-gene-silencing.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
>>> Read full article>>>
Copyright for syndicated content belongs to the linked Source : Phys.org – https://phys.org/news/2024-04-uncovering-key-players-gene-silencing.html










