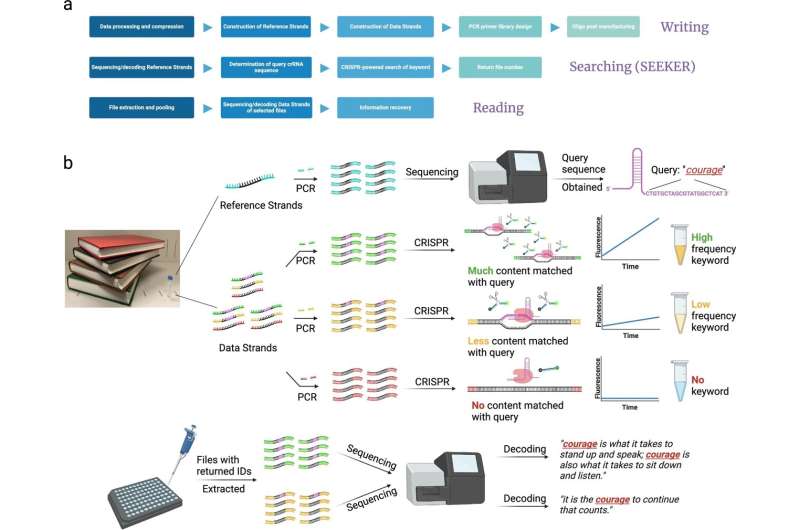
The framework of a DNA data storage system with searching capability and the general workflow of SEEKER. a Complete framework of a searchable DNA data storage system includes writing, searching, and reading the data. b The oligo pool storing text data is separately constructed into two parts: reference strands and data strands. The reference strands usually comprise 100–200 oligos and can be pre-sequenced to determine the dictionary used to map the data strands to binary codes as well as the crRNA spacer sequence of an intended query; for instance, the keyword “courage” corresponds to the sequence “CTGTGCTAGCGTATGGCTCAT” in crRNA. The data strands are selectively amplified according to file IDs and then incubated with the Cas12a-crRNA ribonucleoprotein complex. The fluorescence intensity increases rapidly if the amplified file contains many repeats of the keyword “courage,” generating a strong fluorescence signal within a short time. If fewer instances of the keyword “courage” appear in the file, the fluorescence enhancement retards, and the endpoint fluorescence intensity becomes weaker. If the keyword “courage” is not found in the file, no fluorescence will be detected. After searching, files generating positive signals are recognized as carrying the data of interest and are subjected to next-generation sequencing to recover the complete content. In this example, a stronger signal is generated when the keyword “courage” appears twice, whereas a weaker signal is generated when the keyword only appears once. Illustrations were created with BioRender.com. Credit: Nature Communications (2024). DOI: 10.1038/s41467-024-46767-x
The digital age has led to the explosive growth of data of all kinds. Traditional methods for storing data—such as hard drives—are beginning to face challenges due to limited storage capacities. With the growing demand for data storage on the rise, alternate mediums of data storage are becoming increasingly popular—and necessary.
DNA is one of the emerging solutions to store data due to its physical density, data longevity, and data encryption ability. Any information that can be stored in a hard drive—such as texts, images, sounds, and movies—can also be converted into DNA sequences.
But while DNA is a promising solution to help meet the demand of data storage needs, performing a search within a strand of DNA can be cumbersome and difficult.
“Archiving information in synthetic DNA has emerged as an attractive solution to deal with the exploding growth of data in the modern world. However, quantitatively querying the data stored in DNA is still a challenge,” says Changchun Liu, professor in the Department of Biomedical Engineering at UConn Health.
In Nature Communications, Liu and a team of researchers discovered a way to simply and effectively search for data stored in DNA using a clustered, regularly interspaced short palindromic repeats (CRISPR) powered quantitative search engine.
In the paper, Liu introduces Search Enabled by Enzymatic Keyword Recognition (SEEKER), which utilizes CRISPR-Cas12a to identify the keyword in files stored in DNA quantitatively.
“DNA is a promising medium for data storage because of its stability and high information density. Theoretically, one gram of DNA can store 215 petabytes of data, the data size of about 100 million movies. Like a hard drive that stores information in binary data bits, DNA stores information in sequences of four nucleobases—adenine (A), thymine (T), cytosine (C), and guanine (G).
“The developments in DNA synthesis technology and next-generation sequencing are turning DNA data storage into reality,” explains Jiongyu Zhang, a graduate student in Liu’s lab and first author of the paper.
Liu utilized his expertise in CRISPR technology to help come up with a better solution to search within a strand of DNA.
CRISPR is an acquired immune mechanism that can identify a specific infectious DNA sequence in a cell overwhelmed with interfering genes, similar to a keyword search in a database.
SEEKER, utilizing CRISPR, rapidly generates visible fluorescence, or light, when a DNA target corresponding to the keyword of interest is present. SEEKER is able to successfully perform quantitative text searching since the growth rate of the fluorescence intensity is proportional to the keyword frequency.
In the paper, the researchers successfully identified keywords in 40 files with a background of approximately 8,000 irrelevant terms.
“Overall, the SEEKER provides a quantitative approach to conducting parallel searching-including metadata search—over the complete content stored in DNA with simple implementation and rapid result generation,” explains Liu.
More information:
Jiongyu Zhang et al, CRISPR-powered quantitative keyword search engine in DNA data storage, Nature Communications (2024). DOI: 10.1038/s41467-024-46767-x
Citation:
Searching for data in DNA with CRISPR (2024, March 19)
retrieved 19 March 2024
from https://phys.org/news/2024-03-dna-crispr.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
>>> Read full article>>>
Copyright for syndicated content belongs to the linked Source : Phys.org – https://phys.org/news/2024-03-dna-crispr.html










